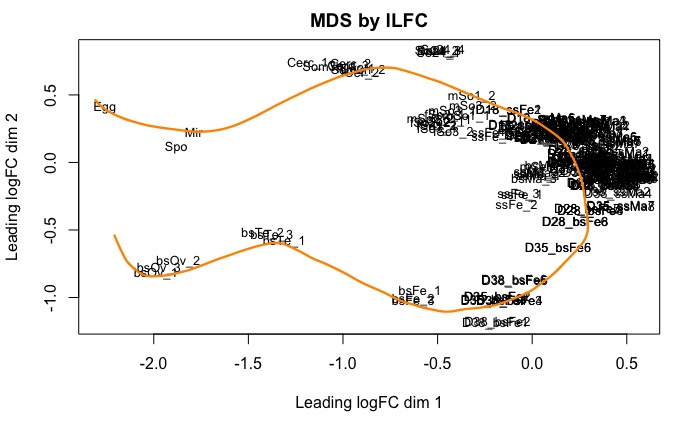
RNA-seq data used for gene expression
RNA-seq data used
Mixed-sex infection
| Group | Label | Sequence Source | Sample Accession | Biological Replicates | Reference |
|---|---|---|---|---|---|
| Gonad (testis) | Te | PRJEB14695 | ERS420096, ERS420097, ERS420098 | 3 | Lu et al. (2016) |
| Gonad (ovary) | Ov | PRJEB14695 | ERS420090, ERS420091, ERS420092 | 3 | Lu et al. (2016) |
| Egg | Egg | PRJNA294789 | SRR2245469 | 1 | Anderson et al. (2015) |
| Miracidium | Mir | PRJNA209511 | SRR922067 | 1 | Wang et al. (2013) |
| Sporocyst (48h) | Spo | PRJNA209511 | SRR922068 | 1 | Wang et al. (2013) |
| Cercaria | Cerc | PRJEB2350 | ERR022872, ERR022877, ERR022878 | 3 | Protasio et al. (2012) |
| Schistosomulum (3h) | Som3h | PRJEB2350 | ERR022874, ERR022876 | 2 | Protasio et al. (2012) |
| Schistosomulum (24h) | Som24h | PRJEB2350 | ERR022880, ERR022881, ERR022882, ERR022883 | 4 | Protasio et al. (2012) |
| Juvenile male 21d | m21d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Juvenile female 21d | f21d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Juvenile male 28d | m28d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Juvenile female 28d | f28d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Adult male 35d | m35d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult female 35d | f35d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult male 38d | m38d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult female 38d | f38d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult male >42d | m42d+ | PRJEB14695 | ERS420093, ERS420106, ERS420107 | 3 | Lu et al. (2016) |
| Adult female >42d | f42d+ | PRJEB14695 | ERS420099, ERS420100, ERS420101 | 3 | Lu et al. (2016) |
Single-sex infection
| Group | Label | Sequence Source | Sample Accession | Biological Replicates | Reference |
|---|---|---|---|---|---|
| Egg | Egg | PRJNA294789 | SRR2245469 | 1 | Anderson et al. (2015) |
| Miracidium | Mir | PRJNA294789 | SRR922067 | 1 | Wang et al. (2013) |
| Sporocyst (48h) | Spo | PRJNA294789 | SRR922068 | 1 | Wang et al. (2013) |
| Cercaria male | mCer | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Cercaria female | fCer | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #1 male | mSo1 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #1 female | fSo1 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #2 male | mSo2 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #2 female | fSo2 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #3 male | mSo3 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Somule stage #3 female | fSo3 | PRJNA312093 | – | 2 | Picard et al. (2016) |
| Juvenile male 18d | m18d | PRJEB3190 | – | 4 | Protasio et al. 2017 |
| Juvenile female 18d | f18d | PRJEB3190 | – | 4 | Protasio et al. 2017 |
| Juvenile male 21d | m21d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Juvenile female 21d | f21d | PRJEB3190 | – | 10 | Protasio et al. 2017 |
| Juvenile male 28d | m28d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Juvenile female 28d | f28d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult male 35d | m35d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Adult female 35d | f35d | PRJEB3190 | – | 6 | Protasio et al. 2017 |
| Adult male 38d | m38d | PRJEB3190 | – | 12 | Protasio et al. 2017 |
| Adult female 38d | f38d | PRJEB3190 | – | 8 | Protasio et al. 2017 |
| Adult male >42d | m42d+ | PRJNA312093 (1), PRJEB14695 (3) | – | 4 | Picard et al. (2016), Lu et al. (2016) |
| Adult female >42d | f42d+ | PRJNA312093 (2), PRJEB14695 (3) | – | 5 | Picard et al. (2016), Lu et al. (2016) |
Note that there are variations in S. mansoni strain and definitive host in these studies.
Analysis pipeline
Reads were mapped to S. mansoni genome annotation v7.2 using HISAT2 (v2.1.0) for egg and STAR (v2.4.2a) for the rest samples. Counts per gene were summarised with featureCounts (v1.4.5-p1) on based on exon features. Read counts were normalised in edgeR (v3.24.3) using the TMM method. RPKM values were calculated based on TMM-normalised counts and mean values were used for biological replicates (indicated as “Normalised expression” in the charts).
Data Exploration